CUDA Parallel Programming/Anova with CUDA
Anova with CUDA
Introduction
Anova can be used to define and solve the mathematical formula of species like Human, Rabbit etc. Each of those species formed by variables like: Height, Eye Color, Bone thickness, Hair Color etc. or sicknesses like Cancer.
As a understanding my problem: Height(It is one of variable that defines Human) is a specific property of each person.
There might be of other variables like eye color; hair color etc. but I will concentrate on one measurement that causing difference that occurs on Gene. Related to my comparison between DNA sequences I have chance to observe different Gene-pairs between each person. By looking my result I have to conclude that which part of SNPs that (different pairs of DNA) causes the height.
Genome-Wide Association Study: is an examination of many common genetic variants in different to see if any variant is associated with a trait. Genetic Variant: A single-nucleotide polymorphism (SNP).
Problem
Compare the DNA of two groups of patients: people worth disease. SNPs are then considered to mark a region of the human genome which influences the risk of disease, eye color, height etc.
My Aim:
I have to find SNP-pairs that have significant association with a given quantitative phenotype (height or weight). ANOVA tests on all SNP pairs Two people, all 3.1 billion molecules of it, is more than 99.9 percent identical but that 0.1 percent accounts for all the genetic differences between people. The difference occurred in people DNA Sequence causes that one person might have blue eyes or lung cancer, or perfect pitch.
Rather than having A-T pair of molecule at a certain spot on the DNA chain, a person might have a G-C pair. On the other hand that difference might now have any effect at all on a person’s health or appearance. These differences also called as SNPs.
For example, if 1000 people share the same disease that all these people share – genetic mutations (SNPs) that healthy people don’t have.
My Data:
DNA sequence of each person. As factor I can change the number of people that I am experience on. I can’t change the original sequence of DNA. Factor to Change: I can change the person that I am using in my dataset. I can compare healthy and not-healthy persons. I can compare tall and short people. By depending on the sequence of DNA each person has its unique characteristics (eye color, height, any diseases …). So in to my experiment data set I can put specific type of person. For example 99 person blue eyes and 1 person as brown eyes, by comparing all of them I will have chance to result that which part of DNA sequence affect eye color as blue.
Example Result
These example runs on functions of ANOVA formula as parallel. It is efficient because data set that contains information related to each object increase CUDA make the implementation faster.
Our data members can be:
int data1_store[MEMBERNUM] = {7, 4, 6, 8, 6, 6, 2, 9 …...};
int data2_store[MEMBERNUM] = {5, 5, 3, 4, 4, 7, 2, 2 …...};
int data3_store[MEMBERNUM] = {2, 4, 7, 1, 2, 1, 5, 5 …...};
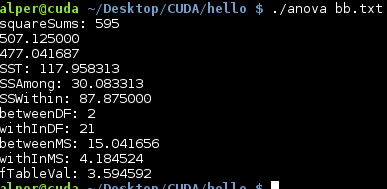
Our result find the relation between each member . By looking F-Table Value we can conclude those members interaction with each other.
Example Source Code
__global__ void squareMemberSum(int *data1,int *data2,int *data3, int *totalSum, int *Res ,int N ){
int idx = threadIdx.x + blockIdx.x * blockDim.x;
int result = 0;
atomicAdd((totalSum + 0), data1[idx] * data1[idx]); atomicAdd((totalSum + 1), data2[idx] * data2[idx]); atomicAdd((totalSum + 2), data3[idx] * data3[idx]);
for(int i = 0; i < GROUPNUM; i++){ result += totalSum[i]; totalSum[i] = 0; } *Res = result; } __global__ void memberSum(int *data1,int *data2,int *data3, int *res , float * R , float *sumSquareT,int N ) { int idx = threadIdx.x + blockIdx.x * blockDim.x;
atomicAdd((res + 0), data1[idx]); atomicAdd((res + 1), data2[idx]); atomicAdd((res + 2), data3[idx]);
for(int i = 0; i < GROUPNUM; i++){ *R += res[i] * res[i]; *sumSquareT += res[i]; }
*R = *R / MEMBERNUM; *sumSquareT = (*sumSquareT) * (*sumSquareT); *sumSquareT = *sumSquareT / (GROUPNUM * MEMBERNUM); }
Measurement:
Compare the DNA sequence of 2 or more than 2 people. Found pair differences are my SNP genes. How many SNP genes there are, where they are located.
My Goal:
To find 0.1 difference of DNA sequence between each person. Depending on my result and looking at my characteristics of each person, I can conclude that which part of DNA sequence causing those characteristics.